|
|
Patricia Marquez, Debora Gil, & Aura Hernandez-Sabate. (2012). "Error Analysis for Lucas-Kanade Based Schemes " In 9th International Conference on Image Analysis and Recognition (Vol. 7324, pp. 184–191). Lecture Notes in Computer Science. Springer-Verlag Berlin Heidelberg.
Abstract: Optical flow is a valuable tool for motion analysis in medical imaging sequences. A reliable application requires determining the accuracy of the computed optical flow. This is a main challenge given the absence of ground truth in medical sequences. This paper presents an error analysis of Lucas-Kanade schemes in terms of intrinsic design errors and numerical stability of the algorithm. Our analysis provides a confidence measure that is naturally correlated to the accuracy of the flow field. Our experiments show the higher predictive value of our confidence measure compared to existing measures.
Keywords: Optical flow, Confidence measure, Lucas-Kanade, Cardiac Magnetic Resonance
|



|
|
|
Aura Hernandez-Sabate, Debora Gil, David Roche, Monica M. S. Matsumoto, & Sergio S. Furuie. (2011). "Inferring the Performance of Medical Imaging Algorithms " In Pedro Real, Daniel Diaz-Pernil, Helena Molina-Abril, Ainhoa Berciano, & Walter Kropatsch (Eds.), 14th International Conference on Computer Analysis of Images and Patterns (Vol. 6854, pp. 520–528). L. Berlin: Springer-Verlag Berlin Heidelberg.

Abstract: Evaluation of the performance and limitations of medical imaging algorithms is essential to estimate their impact in social, economic or clinical aspects. However, validation of medical imaging techniques is a challenging task due to the variety of imaging and clinical problems involved, as well as, the difficulties for systematically extracting a reliable solely ground truth. Although specific validation protocols are reported in any medical imaging paper, there are still two major concerns: definition of standardized methodologies transversal to all problems and generalization of conclusions to the whole clinical data set.
We claim that both issues would be fully solved if we had a statistical model relating ground truth and the output of computational imaging techniques. Such a statistical model could conclude to what extent the algorithm behaves like the ground truth from the analysis of a sampling of the validation data set. We present a statistical inference framework reporting the agreement and describing the relationship of two quantities. We show its transversality by applying it to validation of two different tasks: contour segmentation and landmark correspondence.
Keywords: Validation, Statistical Inference, Medical Imaging Algorithms.
|



|
|
|
Aura Hernandez-Sabate, Petia Radeva, Antonio Tovar, & Debora Gil. (2006). "Vessel structures alignment by spectral analysis of ivus sequences " In Proc. of CVII, MICCAI Workshop (pp. 39–36). 1st International Wokshop on Computer Vision for Intravascular and Intracardiac Imaging (CVII’06). Copenhaguen (Denmark),.

Abstract: Three-dimensional intravascular ultrasound (IVUS) allows to visualize and obtain volumetric measurements of coronary lesions through an exploration of the cross sections and longitudinal views of arteries. However, the visualization and subsequent morpho-geometric measurements in IVUS longitudinal cuts are subject to distortion caused by periodic image/vessel motion around the IVUS catheter. Usually, to overcome the image motion artifact ECG-gating and image-gated approaches are proposed, leading to slowing the pullback acquisition or disregarding part of IVUS data. In this paper, we argue that the image motion is due to 3-D vessel geometry as well as cardiac dynamics, and propose a dynamic model based on the tracking of an elliptical vessel approximation to recover the rigid transformation and align IVUS images without loosing any IVUS data. We report an extensive validation with synthetic simulated data and in vivo IVUS sequences of 30 patients achieving an average reduction of the image artifact of 97% in synthetic data and 79% in real-data. Our study shows that IVUS alignment improves longitudinal analysis of the IVUS data and is a necessary step towards accurate reconstruction and volumetric measurements of 3-D IVUS.
|



|
|
|
Joel Barajas, Jaume Garcia, Francesc Carreras, Sandra Pujades, & Petia Radeva. (2005). "Angle Images Using Gabor Filters in Cardiac Tagged MRI " In Proceeding of the 2005 conference on Artificial Intelligence Research and Development (pp. 107–114). Amsterdam, The Netherlands: IOS Press.
Abstract: Tagged Magnetic Resonance Imaging (MRI) is a non-invasive technique used to examine cardiac deformation in vivo. An Angle Image is a representation of a Tagged MRI which recovers the relative position of the tissue respect to the distorted tags. Thus cardiac deformation can be estimated. This paper describes a new approach to generate Angle Images using a bank of Gabor filters in short axis cardiac Tagged MRI. Our method improves the Angle Images obtained by global techniques, like HARP, with a local frequency analysis. We propose to use the phase response of a combination of a Gabor filters bank, and use it to find a more precise deformation of the left ventricle. We demonstrate the accuracy of our method over HARP by several experimental results.
Keywords: Angle Images, Gabor Filters, Harp, Tagged Mri
|



|
|
|
Jaume Garcia, Joel Barajas, Francesc Carreras, Sandra Pujades, & Petia Radeva. (2005). "An intuitive validation technique to compare local versus global tagged MRI analysis " In Computers In Cardiology (Vol. 32, 29–32).
Abstract: Myocardium appears as a uniform tissue that seen in convectional Magnetic Resonance Images (MRI) shows just the contractile part of its movement. MR Tagging is a unique imaging technique that prints a grid over the tissue which moves according to the underlying movement of the myocardium revealing the true deformation of the cardiac muscle. Optical flow techniques based on spectral information estimate tissue displacement by analyzing information encoded in the phase maps which can be obtained using, local (Gabor) and global (HARP) methods. In this paper we compare both in synthetic and real Tagged MR sequences. We conclude that local method is slightly more accurate than the global one. On the other hand, global method is more efficient as it is much faster and less parameters have to be taken into account
|



|
|
|
Debora Gil, Jaume Garcia, Mariano Vazquez, Ruth Aris, & Guilleaume Houzeaux. (2008). "Patient-Sensitive Anatomic and Functional 3D Model of the Left Ventricle Function " In 8th World Congress on Computational Mechanichs (WCCM8).

Abstract: Early diagnosis and accurate treatment of Left Ventricle (LV) dysfunction significantly increases the patient survival. Impairment of LV contractility due to cardiovascular diseases is reflected in its motion patterns. Recent advances in medical imaging, such as Magnetic Resonance (MR), have encouraged research on 3D simulation and modelling of the LV dynamics. Most of the existing 3D models [1] consider just the gross anatomy of the LV and restore a truncated ellipse which deforms along the cardiac cycle. The contraction mechanics of any muscle strongly depends on the spatial orientation of its muscular fibers since the motion that the muscle undergoes mainly takes place along the fibers. It follows that such simplified models do not allow evaluation of the heart electro-mechanical function and coupling, which has recently risen as the key point for understanding the LV functionality [2]. In order to thoroughly understand the LV mechanics it is necessary to consider the complete anatomy of the LV given by the orientation of the myocardial fibres in 3D space as described by Torrent Guasp [3].
We propose developing a 3D patient-sensitive model of the LV integrating, for the first time, the ven- tricular band anatomy (fibers orientation), the LV gross anatomy and its functionality. Such model will represent the LV function as a natural consequence of its own ventricular band anatomy. This might be decisive in restoring a proper LV contraction in patients undergoing pace marker treatment.
The LV function is defined as soon as the propagation of the contractile electromechanical pulse has been modelled. In our experiments we have used the wave equation for the propagation of the electric pulse. The electromechanical wave moves on the myocardial surface and should have a conductivity tensor oriented along the muscular fibers. Thus, whatever mathematical model for electric pulse propa- gation [4] we consider, the complete anatomy of the LV should be extracted.
The LV gross anatomy is obtained by processing multi slice MR images recorded for each patient. Information about the myocardial fibers distribution can only be extracted by Diffusion Tensor Imag- ing (DTI), which can not provide in vivo information for each patient. As a first approach, we have
Figure 1: Scheme for the Left Ventricle Patient-Sensitive Model.
computed an average model of fibers from several DTI studies of canine hearts. This rough anatomy is the input for our electro-mechanical propagation model simulating LV dynamics. The average fiber orientation is updated until the simulated LV motion agrees with the experimental evidence provided by the LV motion observed in tagged MR (TMR) sequences. Experimental LV motion is recovered by applying image processing, differential geometry and interpolation techniques to 2D TMR slices [5]. The pipeline in figure 1 outlines the interaction between simulations and experimental data leading to our patient-tailored model.
Keywords: Left Ventricle, Electromechanical Models, Image Processing, Magnetic Resonance.
|


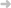
|
|
|
C. Santa-Marta, Jaume Garcia, A. Bajo, J.J. Vaquero, M. Ledesma-Carbayo, & Debora Gil. (2008)." Influence of the Temporal Resolution on the Quantification of Displacement Fields in Cardiac Magnetic Resonance Tagged Images" In S. A. Roberto hornero (Ed.), XXVI Congreso Anual de la Sociedad Española de Ingenieria Biomedica (352–353).
Abstract: It is difficult to acquire tagged cardiac MR images with a high temporal and spatial resolution using clinical MR scanners. However, if such images are used for quantifying scores based on motion, it is essential a resolution as high as possibl e. This paper explores the influence of the temporal resolution of a tagged series on the quantification of myocardial dynamic parameters. To such purpose we have designed a SPAMM (Spatial Modulation of Magnetization) sequence allowing acquisition of sequences at simple and double temporal resolution. Sequences are processed to compute myocardial motion by an automatic technique based on the tracking of the harmonic phase of tagged images (the Harmonic Phase Flow, HPF). The results have been compared to manual tracking of myocardial tags. The error in displacement fields for double resolution sequences reduces 17%.
|

|
|
|
Joel Barajas, Jaume Garcia, Karla Lizbeth Caballero, Francesc Carreras, Sandra Pujades, & Petia Radeva. (2006). "Correction of Misalignment Artifacts Among 2-D Cardiac MR Images in 3-D Space " In 1st International Wokshop on Computer Vision for Intravascular and Intracardiac Imaging (CVII’06) (Vol. 3217, pp. 114–121). Copenhagen (Denmark).
Abstract: Cardiac Magnetic Resonance images offer the opportunity to study the heart in detail. One of the main issues in its modelling is to create an accurate 3-D reconstruction of the left ventricle from 2-D views. A first step to achieve this goal is the correct registration among the different image planes due to patient movements. In this article, we present an accurate method to correct displacement artifacts using the Normalized Mutual Information. Here, the image views are treated as planes in order to diminish the approximation error caused by the association of a certain thickness, and moved simultaneously to avoid any kind of bias in the alignment process. This method has been validated using real and syntectic plane displacements, yielding promising results.
|


|
|
|
Cristina Cañero, Petia Radeva, Oriol Pujol, Ricardo Toledo, Debora Gil, J. Saludes, et al. (1999). "Optimal Stent Implantation: Three-dimensional Evaluation of the Mutual Position of Stent and Vessel via Intracoronary Ecography " In Proceedings of International Conference on Computer in Cardiology (CIC´99).
Abstract: We present a new automatic technique to visualize and quantify the mutual position between the stent and the vessel wall by considering their three-dimensional reconstruction. Two deformable generalized cylinders adapt to the image features in all IVUS planes corresponding to the vessel wall and the stent in order to reconstruct the boundaries of the stent and the vessel in space. The image features that characterize the stent and the vessel wall are determined in terms of edge and ridge image detectors taking into account the gray level of the image pixels. We show that the 30 reconstruction by deformable cylinders is accurate and robust due to the spatial data coherence in the considered volumetric IVUS image. The main clinic utility of the stent and vessel reconstruction by deformable’ cylinders consists of its possibility to visualize and to assess the optimal stent introduction.
|


|
|
|
Cristina Cañero, Petia Radeva, Oriol Pujol, Ricardo Toledo, Debora Gil, J. Saludes, et al. (1999). "Three-dimensional reconstruction and quantification of the coronary tree using intravascular ultrasound images " In Proceedings of International Conference on Computer in Cardiology (CIC´99).
Abstract: In this paper we propose a new Computer Vision technique to reconstruct the vascular wall in space using a deformable model-based technique and compounding methods, based in biplane angiography and intravascular ultrasound data jicsion. It is also proposed a generalpurpose three-dimensional guided interpolation method. The three dimensional centerline of the vessel is reconstructed from geometrically corrected biplane angiographies using automatic segmentation methods and snakes. The IVUS image planes are located in the threedimensional space and correctly oriented. A led interpolation method based in B-SurJaces and snakes isused to fill the gaps among image planes
|


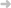
|